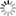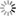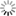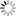Neurogenomics meets bioinformatics meets neuroinformatics
Chair: Robert Williams, University of Tennessee Health Science Center, USA
 Massive data sets on the expression of genes and proteins in the central nervous systems are rapidly altering the neuroscience research landscape and are fundamentally changing how investigators develop and test hypotheses.
Massive data sets on the expression of genes and proteins in the central nervous systems are rapidly altering the neuroscience research landscape and are fundamentally changing how investigators develop and test hypotheses.
This workshop will highlight powerful and open genomic resources that are part of a new wave of information dissemination. Examples include the Allen Brain Atlas, GeneNetwork, and Genes2Cognition. The workshop leaders will illustrate how neurogenomic resources are having an impact on our basic understanding of brain structure and function. They will also illustrate how these resources are having an impact on translational research. Exploiting neurogenomic resources usually requires judicious combination with large data sets on DNA, RNA, and protein sequence and variation. One of the major goals of neuroinformatics is to successfully blend these highly diverse bioinformatic resources with the rapidly growing neuroscience literature.
 It should soon be possible to efficiently and correctly answer specific questions about brain function and disease using sophisticated neuroinformatic web portals such as that being built by the INCF and BIRN. This entails the assembly of distributed computer systems that are coupled by well established conventions for data annotation, calculation, and information exchange.
It should soon be possible to efficiently and correctly answer specific questions about brain function and disease using sophisticated neuroinformatic web portals such as that being built by the INCF and BIRN. This entails the assembly of distributed computer systems that are coupled by well established conventions for data annotation, calculation, and information exchange.
Invited speakers:
Allen Institute for Brain Science, Seattle, USA
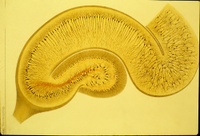
Title: “Genomic Neuroanatomy”: Novel principles of hippocampal organization gained from analysis of genome-scale in situ hybridization data

Abstract: Differential gene expression underlies differential cellular phenotype in the nervous system, and individual gene expression patterns have long been used to delineate functionally discrete neuroanatomical subdivisions. This type of approach to define the genetic architecture, or “molecular anatomy,” of the brain has been radically bolstered by the recent availability of genome-scale cellular resolution in situ hybridization data in the Allen Brain Atlas and related databases. In particular, the mapping of these data to standardized anatomical models allows statistical analyses across these large data sets to understand genetic regional parcellation and interrelationships. In the current study, a variety of statistical clustering methods and classical anatomical techniques were used to examine the genetic architecture of the adult mouse hippocampus across the entire Allen Brain Atlas data set. These data demonstrate a robust genetic correlate to the classical anatomical parcellation of the hippocampus into dentate gyrus, CA1, CA2 and CA3 fields, but also an unanticipated degree of molecular heterogeneity along the septotemporal axis of the hippocampus as well. While individual gene expression patterns are highly diverse, as a set these data show a coherent, complex cellular organization represented by unique combinatorial gene expression patterns highly representing functionally relevant gene families such as transcription factors, ion channels and cell adhesion molecules. This approach to “genomic neuroanatomy” provides both high level organizational principles and a high resolution map of gene expression profiles to guide functional studies, and should be a powerful strategy to expand upon as more large-scale gene expression data sets become available.
Wellcome Trust Sanger Institute, Cambridge, UK
Title: Synapse systems biology
Abstract: Proteomic studies show that mammalian synapses comprise 1-2000 proteins organised into networks and multiprotein complexes. There is an important need to understand the logic behind the complexity of synapses. Toward this objective we have mapped and integrated protein interactions, phosphorylation, mRNA and protein expression profiles and phenotypic maps based on mutations of the synapse,. These maps show a highly integrated signaling machine in the postsynaptic proteome with an extraordinary number of combinations of functional states. We have recently completed a study on the molecular evolution of synapses and find that an ancestral protosynapse derived from core proteins was embellished during evolution to produce the complexity observed in mammals. This has lead to a general theory on the origins of synapse complexity and its role in species differences and complex behaviors.
University of Texas at Austin, USA
Title: Using Neuroinformatics Tools to investigate and share high resolution full volume recontructions of brain neuropil 
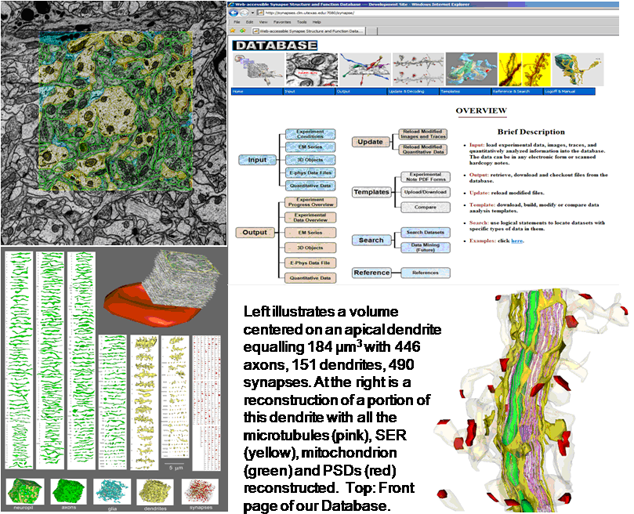
Abstract: Full volume manual reconstructions have been obtained to ascertain the synaptic connectivity and composition of the neuropil surrounding individual hippocampal CA1 dendrites and dendritic spines in mature hippocampus. All objects in two cylindrical volumes were manually reconstructed. These and other test volumes are being used to evaluate the accuracy of automated segmentation algorithms and expand analysis beyond the manual-reconstructions to other dendrites in these neuropils. The resolution of ssTEM provides unambiguous identification of synapses and the subcellular composition of organelles in dendrites, axons, and glia. Thus, full volume high resolution reconstruction provides a powerful tool to determine synaptic circuitry and relate it specifically to alterations in the composition of the local neuropil and small subcellular organelles, such as polyribosomes, vesicles, and glycogen granules, which can serve to identify the local functional status of dendrites, axons, and glia. We also hypothesize that the capacity for synaptic plasticity is dependent on the local availability of core subcellular structures (such as mitochondria (green), vesicles, polyribosomes, mRNA, SER (yellow), Golgi apparatus) that help to build and maintain synapses. Transport along microtubules is required to deliver the core structures. Hence, we used reconstruction from ssTEM to test whether dendritic spine density (#/µm) was correlated with the number of microtubules in a dendritic branch. We are developing a neuroinformatics database of the images and traces comprising these full-volume and subcellular reconstructions to share with broader community, both to develop additional reconstruction and analytical tools, and to model local dendritic and axonal connectivity and function.